II类V型CRISPR蛋白CeCas12a及其在基因编辑的应用的制作方法
本发明属于生物医学领域,具体涉及一种来自于coprococcuseutactus细菌中的ii类v型crispr蛋白cas12a,命名为cecas12a,在基因编辑中的应用。
背景技术:
自2013年以来,基因编辑技术取得了突破性进展,此项技术已经在基础科学研究、医药、临床、生物技术等许多领域引起了新的变革。除了具有代表性的cas9系统之外,cas12,又名cpf1,作为又一种被发现的具有基因编辑效应的crispr系统新成员,极大的扩大了基因编辑系统靶点的可编辑范围,相比于cas9系统,cas12a所具有的加工前提rna的功能,为其介导多基因编辑提供了相比与cas9系统更为便捷高效的编辑能力。除此之外,相比于cas9的向导rna,cas12a的向导rna组成更为简单,设计更为方便。
2015年,张峰团队首次发现了cas9系统之外的另外一种具有基因编辑能力的新成员,cas12a,又名cpf1,将其划分到crispr系统2类v型中。相比于cas9系统,cas12a的编辑效率与cas9的效率相当,在有些靶点低于cas9。cas12a的脱靶率极低,相比于cas9脱靶率高的特性,cas12a是一种安全的基因编辑工具。cas12a在切割之后形成粘性末端,而cas9形成平末端,已有研究表明,cas12a切割之后的粘性末端相比于cas9的平末端而言,更容易发生同源重组修复,这也为基因的定点插入和修复提供了更好的工具。在向导rna的加工方面,cas12a具有明显的优势,仅仅只需要cas12a本身就能够完成对前提rna的加工,而cas9系统则需要rnaseiii的加工,这极大地促进cas12a在多基因编辑上的应用。在pam的识别上,cas12a识别5’-tttn-3’或5’-kytv-3’,cas9则识别5’-ngg-3’。
因此,cas12a作为一种新型基因编辑工具,与cas9系统一道,为科学研究和疾病的治疗提供了有力的工具。基于对目前已有的cas12a的研究,为应对将来各种情况下的基因编辑事件,发现更多的具有一定特性的cas12a是一件具有重要意义的事情。
技术实现要素:
本发明针对现有技术的不足,目的在于提供一种来自于coprococcuseutactus细菌中的ii类v型crispr蛋白cecas12a及其在基因编辑中的应用。
为实现上述发明目的,本发明所采用的技术方案为:
一种来自于coprococcuseutactus细菌中的ii类v型crispr蛋白cecas12a,其氨基酸序列如seqidno.1所示。
上述方案中,所述cecas12a识别的pam序列为tttv、tcta、ttca、或ctta,更为优选的pam序列为tttv,所述v表示a、c、或g。
用于编辑蛋白cecas12a氨基酸序列的基因,其核苷酸序列如seqidno.2所示。
上述cecas12a在基因编辑中的应用。
上述cecas12a在原核生物基因编辑中的应用。
上述cecas12a在真核生物基因编辑中的应用。
上述cecas12a在体外基因编辑中的应用。
本发明所述蛋白cecas12a的氨基酸序列如下:
(1)原核细胞中:
nnntnnsfepfiggnsvsktlrnelrvgseytgkhikecaiiaedavkaenqyivkemmddfyrdfinrkldalqginweqlfdimkkakldksnkvskeldkiqestrkeivkifssdpiykdmlkadmiskilpeyivdkygdaasrieavkvfygfsgyfidfwasrknvfsdkniasaiphrivnvnarihldnitafnriaeiagdevagiaedacaylqnmsledvftgacygeficqkdidrynnicgvinqhmnqycqnkkisrskfkmerlhkqilcrsesgfeipigfqtdgevidainsfstileekdildrlrtlsqevtgydmeriyvsskafesvskyidhkwdviassmynyfsgavrgkddkkdakiqteikkikscslldlkklvdmyykmdgmcleheateyvagiteilvdfnyktfdmddsvkmiqnehmineikeyldtymsiyhwakdfmidelvdrdmefyseldeiyydlsdivplynkvrnyvtqkpysqdkiklnfgsptlangwskskefdnnvvvllrdekiylailnvgnkpskdimagedrrrsdtdykkmnyyllpgasktlphvfissnawkkshgipdeimygynqnkhlksspnfdlefcrklidyykecidsypnyqifnfkfaatetyndisefykdverqgykiewsyiseddinqmdrdgqiylfqiynkdfapnskgmqnlhtlylknifseenlsdvviklngeaelffrkssiqhkrghkkgsvlvnktykttektengqgeieviesvpdqcylelvkywseggvgqlseeaskykdkvshyaatmdivkdrrytedkffihmpitinfkadnrnnvnekvlkfiaenddlhvigidrgernllyvsvidsrgriveqksfnivenyessknvirrhdykgklvnkehyrnearkswkeigkikeikegylsqviheisklvlkynaiivmedlnygfkrgrfkverqvyqkfetmlinklaylvdksravdepggllkgyqltyvpdnlgelgsqcgiifyvpaaytskidpvtgfvdvfdfkaysnaearldfinkldcirydasrnkfeiafdygnfrthhttlaktswtifihgdrikkergsygwkdeiidiearirklfedtdieyadghnligdinelespiqkkfvgelfdiirftvqlrnsksekydgtekeydkiispvmdeegvffttdsyiradgtelpkdadangaycialkglydvlavkkywkegekfdrkllaitnynwfdfiqnrrf
(2)真核细胞中:
pkkkrkvnnntnnsfepfiggnsvsktlrnelrvgseytgkhikecaiiaedavkaenqyivkemmddfyrdfinrkldalqginweqlfdimkkakldksnkvskeldkiqestrkeivkifssdpiykdmlkadmiskilpeyivdkygdaasrieavkvfygfsgyfidfwasrknvfsdkniasaiphrivnvnarihldnitafnriaeiagdevagiaedacaylqnmsledvftgacygeficqkdidrynnicgvinqhmnqycqnkkisrskfkmerlhkqilcrsesgfeipigfqtdgevidainsfstileekdildrlrtlsqevtgydmeriyvsskafesvskyidhkwdviassmynyfsgavrgkddkkdakiqteikkikscslldlkklvdmyykmdgmcleheateyvagiteilvdfnyktfdmddsvkmiqnehmineikeyldtymsiyhwakdfmidelvdrdmefyseldeiyydlsdivplynkvrnyvtqkpysqdkiklnfgsptlangwskskefdnnvvvllrdekiylailnvgnkpskdimagedrrrsdtdykkmnyyllpgasktlphvfissnawkkshgipdeimygynqnkhlksspnfdlefcrklidyykecidsypnyqifnfkfaatetyndisefykdverqgykiewsyiseddinqmdrdgqiylfqiynkdfapnskgmqnlhtlylknifseenlsdvviklngeaelffrkssiqhkrghkkgsvlvnktykttektengqgeieviesvpdqcylelvkywseggvgqlseeaskykdkvshyaatmdivkdrrytedkffihmpitinfkadnrnnvnekvlkfiaenddlhvigidrgernllyvsvidsrgriveqksfnivenyessknvirrhdykgklvnkehyrnearkswkeigkikeikegylsqviheisklvlkynaiivmedlnygfkrgrfkverqvyqkfetmlinklaylvdksravdepggllkgyqltyvpdnlgelgsqcgiifyvpaaytskidpvtgfvdvfdfkaysnaearldfinkldcirydasrnkfeiafdygnfrthhttlaktswtifihgdrikkergsygwkdeiidiearirklfedtdieyadghnligdinelespiqkkfvgelfdiirftvqlrnsksekydgtekeydkiispvmdeegvffttdsyiradgtelpkdadangaycialkglydvlavkkywkegekfdrkllaitnynwfdfiqnrrfkrpaatkkagqakkkkgsypydvpdyaypydvpdyaypydvpdya
其中在cecas12a蛋白氨基酸序列的n端加入pkkkrkv序列(该序列为n端nls入核序列),在cecas12a蛋白氨基酸序列的c端加入krpaatkkagqakkkk序列(该序列为c端nls入核序列),随后用gs序列连接ypydvpdyaypydvpdyaypydvpdya序列(该序列为3ha序列)。
编码本发明所述蛋白cecas12a的核苷酸序列如下:
aacaacaacaccaacaacagcttcgagcccttcatcggcggcaacagcgtgagcaagaccctgcgcaacgagctgcgcgtgggcagcgagtacaccggcaagcacatcaaggagtgcgccatcatcgccgaggacgccgtgaaggccgagaaccagtacatcgtgaaggagatgatggacgacttctaccgcgacttcatcaaccgcaagctggacgccctgcagggcatcaactgggagcagctgttcgacatcatgaagaaggccaagctggacaagagcaacaaggtgagcaaggagctggacaagatccaggagagcacccgcaaggagatcgtgaagatcttcagcagcgaccccatctacaaggacatgctgaaggccgacatgatcagcaagatcctgcccgagtacatcgtggacaagtacggcgacgccgccagccgcatcgaggccgtgaaggtgttctacggcttcagcggctacttcatcgacttctgggccagccgcaagaacgtgttcagcgacaagaacatcgccagcgccatcccccaccgcatcgtgaacgtgaacgcccgcatccacctggacaacatcaccgccttcaaccgcatcgccgagatcgccggcgacgaggtggccggcatcgccgaggacgcctgcgcctacctgcagaacatgagcctggaggacgtgttcaccggcgcctgctacggcgagttcatctgccagaaggacatcgaccgctacaacaacatctgcggcgtgatcaaccagcacatgaaccagtactgccagaacaagaagatcagccgcagcaagttcaagatggagcgcctgcacaagcagatcctgtgccgcagcgagagcggcttcgagatccccatcggcttccagaccgacggcgaggtgatcgacgccatcaacagcttcagcaccatcctggaggagaaggacatcctggaccgcctgcgcaccctgagccaggaggtgaccggctacgacatggagcgcatctacgtgagcagcaaggccttcgagagcgtgagcaagtacatcgaccacaagtgggacgtgatcgccagcagcatgtacaactacttcagcggcgccgtgcgcggcaaggacgacaagaaggacgccaagatccagaccgagatcaagaagatcaagagctgcagcctgctggacctgaagaagctggtggacatgtactacaagatggacggcatgtgcctggagcacgaggccaccgagtacgtggccggcatcaccgagatcctggtggacttcaactacaagaccttcgacatggacgacagcgtgaagatgatccagaacgagcacatgatcaacgagatcaaggagtacctggacacctacatgagcatctaccactgggccaaggacttcatgatcgacgagctggtggaccgcgacatggagttctacagcgagctggacgagatctactacgacctgagcgacatcgtgcccctgtacaacaaggtgcgcaactacgtgacccagaagccctacagccaggacaagatcaagctgaacttcggcagccccaccctggccaacggctggagcaagagcaaggagttcgacaacaacgtggtggtgctgctgcgcgacgagaagatctacctggccatcctgaacgtgggcaacaagcccagcaaggacatcatggccggcgaggaccgccgccgcagcgacaccgactacaagaagatgaactactacctgctgcccggcgccagcaagaccctgccccacgtgttcatcagcagcaacgcctggaagaagagccacggcatccccgacgagatcatgtacggctacaaccagaacaagcacctgaagagcagccccaacttcgacctggagttctgccgcaagctgatcgactactacaaggagtgcatcgacagctaccccaactaccagatcttcaacttcaagttcgccgccaccgagacctacaacgacatcagcgagttctacaaggacgtggagcgccagggctacaagatcgagtggagctacatcagcgaggacgacatcaaccagatggaccgcgacggccagatctacctgttccagatctacaacaaggacttcgcccccaacagcaagggcatgcagaacctgcacaccctgtacctgaagaacatcttcagcgaggagaacctgagcgacgtggtgatcaagctgaacggcgaggccgagctgttcttccgcaagagcagcatccagcacaagcgcggccacaagaagggcagcgtgctggtgaacaagacctacaagaccaccgagaagaccgagaacggccagggcgagatcgaggtgatcgagagcgtgcccgaccagtgctacctggagctggtgaagtactggagcgagggcggcgtgggccagctgagcgaggaggccagcaagtacaaggacaaggtgagccactacgccgccaccatggacatcgtgaaggaccgccgctacaccgaggacaagttcttcatccacatgcccatcaccatcaacttcaaggccgacaaccgcaacaacgtgaacgagaaggtgctgaagttcatcgccgagaacgacgacctgcacgtgatcggcatcgaccgcggcgagcgcaacctgctgtacgtgagcgtgatcgacagccgcggccgcatcgtggagcagaagagcttcaacatcgtggagaactacgagagcagcaagaacgtgatccgccgccacgactacaagggcaagctggtgaacaaggagcactaccgcaacgaggcccgcaagagctggaaggagatcggcaagatcaaggagatcaaggagggctacctgagccaggtgatccacgagatcagcaagctggtgctgaagtacaacgccatcatcgtgatggaggacctgaactacggcttcaagcgcggccgcttcaaggtggagcgccaggtgtaccagaagttcgagaccatgctgatcaacaagctggcctacctggtggacaagagccgcgccgtggacgagcccggcggcctgctgaagggctaccagctgacctacgtgcccgacaacctgggcgagctgggcagccagtgcggcatcatcttctacgtgcccgccgcctacaccagcaagatcgaccccgtgaccggcttcgtggacgtgttcgacttcaaggcctacagcaacgccgaggcccgcctggacttcatcaacaagctggactgcatccgctacgacgccagccgcaacaagttcgagatcgccttcgactacggcaacttccgcacccaccacaccaccctggccaagaccagctggaccatcttcatccacggcgaccgcatcaagaaggagcgcggcagctacggctggaaggacgagatcatcgacatcgaggcccgcatccgcaagctgttcgaggacaccgacatcgagtacgccgacggccacaacctgatcggcgacatcaacgagctggagagccccatccagaagaagttcgtgggcgagctgttcgacatcatccgcttcaccgtgcagctgcgcaacagcaagagcgagaagtacgacggcaccgagaaggagtacgacaagatcatcagccccgtgatggacgaggagggcgtgttcttcaccaccgacagctacatccgcgccgacggcaccgagctgcccaaggacgccgacgccaacggcgcctactgcatcgccctgaagggcctgtacgacgtgctggccgtgaagaagtactggaaggagggcgagaagttcgaccgcaagctgctggccatcaccaactacaactggttcgacttcatccagaaccgccgcttc
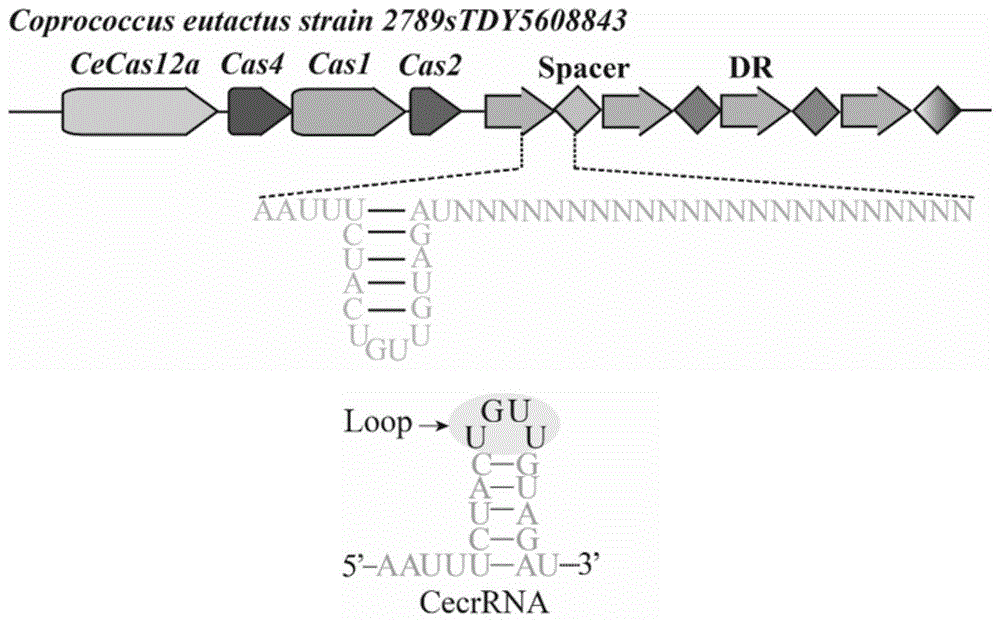
本发明中coprococcuseutactus菌株基因组中部分crisprarray如图2所示,基因组中黑体显示crisprarray序列为“atctacaacagtagaaattatctataggttcttgg”,因此,使用的crrnadirectrepeat序列为5’-aatttctactgttgtagat-3’。
本发明的有益效果:本发明首次在coprococcuseutactus菌株中鉴定出具有基因编辑效应的ii类v型crispr蛋白,命名为cecas12a;所述cecas12a能够在crrna的介导下定点对原核生物和真核生物基因组进行基因编辑,cecas12a的发现进一步扩大了基因编辑工具的种类,同时也为后续各种不同情况的基因编辑提供了重要的备选工具,对基础科研和临床治疗具有十分重要的作用。
附图说明
图1为coprococcuseutactus菌株crisprarray及crrnadirectrepeat图示。
图2为coprococcuseutactus菌株基因组中存在的部分crisprarray示意图。
图3为体外切割egfp片段靶点示意图。
图4为原核表达cecas12a之后,体外切割实验,s表示substrate;p表示product。
图5为体外实验验证cecas12a的pam,s表示substrate;p表示product。
图6为体内验证cecas12a基因编辑,s表示substrate;p表示product。
图7为体内验证cecas12a利用加工前提crrna来同时编辑多个基因。
图8为深度测序验证cecas12a的脱靶率。
具体实施方式
为了更好地理解本发明,下面结合实施例进一步阐明本发明的内容,但本发明的内容不仅仅局限于下面的实施例。
实施例1
cecas12a体外不同时间梯度切割实验,包括以下实验步骤:
(1)cecas12a蛋白的表达与纯化:将cecas12a基因序列合成到pet28a表达载体上,且在c末端加上6his标签,随后将合成好的质粒转化到e.colirosseta2(de3)表达菌株中,挑取单克隆,小量表达检测确定蛋白表达后进行蛋白的大量表达与纯化;重组蛋白依次经过ni柱亲和层析,heparin柱层析,superdex200分子筛纯化后,保存在buffer(10mmtris-hcl,200mmnacl,1mmmgcl)中,并冻存于-80℃备用;
(2)使用crrnadirectrepeat序列为:5’-aatttctactgttgtagat-3’,在体外转录获得crrna;将步骤(1)所得cecas12a蛋白与crrna混合得到cecas12a-crrna复合物;
(3)取100nmcecas12a-crrna复合物与300ng线性化的底物(图4所示)混匀,37℃孵育分别孵育0,1,2,5,10min后,加入适量蛋白酶k,58℃消化60min,跑2%琼脂糖胶,结果如图4所示,cecas12a具有良好的体外切割能力。
实施例2
cecas12a识别pam的确定
(1)设计nnnn四个位置随机组合的上下游引物(n表示a、g、c、t),以egfp片段作为模板,采用overlappcr方法进行pcr,得到256种带有不同pam序列,但spacer序列一样的1.1kb的线性化底物;
(2)取100nmcecas12a-crrna复合物与300ng线性化的底物混匀,37℃孵育分别孵育10min后,加入适量蛋白酶k,58℃消化60min,跑2%琼脂糖胶,部分结果如图5所示,cecas12a能够识别不同的pam:ttta、tcta、ttca、ctta,但最优pam为tttv(v表示a,c,g)。
实施例3
cecas12a在哺乳动物细胞内不同基因的编辑:
(1)构建cecas12a真核表达质粒:将cecas12a基因序列合成到pet28a表达载体上构建cecas12a真核表达质粒;
(2)在哺乳动物细胞中,以293t细胞为例选择trac、trbc、b2m、ctla4、pd1五个基因,分别以这五个基因为目标,构建5个由u6启动子启动转录的u6-crrnaspacer真核表达质粒;
(3)分别针对这五个基因的切割靶点附近设计surveyorprimer,并验证pcr引物的特异性;
(4)消化293t细胞,适当浓度铺24孔板,每孔500ul;
(5)24孔板共转cecas12a真核表达质粒(700ng)和u6-crrnaspacer真核表达质粒(300ng),48h后裂解细胞,取1ul裂解液作为模板、以步骤(3)设计的surveyorprimer引物进行pcr,纯化pcr产物;
(6)取300ngpcr产物与1ul10xt7eibuffer混匀,按以下pcr程序进行复性95℃10min,95℃至85℃-2℃/s,85℃到25℃-0.25℃/s,25℃持续1min,复性之后产物加入1ult7ei,37℃酶切20min,跑2%琼脂糖胶,结果如图6所示在trac、trbc、b2m、ctla4、pd1能够进行基因编辑,编辑效率分别为12%,18%,31%,26%,19%(本案例中的四个基因只是作为代表进行列举)。
实施例4
cecas12a在哺乳动物细胞内进行多基因编辑:
(1)选取并设计由一个u6启动子启动转录形成pre-crrnaarray,pre-crrna为同时包括trac、trbc、b2m、ctla4四个基因的crrna序列;
(2)切割靶点附近设计surveyorprimer,并验证pcr引物的特异性;
(3)消化293t细胞,适当浓度铺24孔板,每孔500ul;
(4)24孔板共转cecas12a真核表达质粒(700ng)和步骤(1)获得的包含trac、trbc、b2m、ctla4四个基因的u6-crrnaspacer真核表达质粒(300ng),48h后裂解细胞,取1ul裂解液作为模板、以步骤(3)设计的surveyorprimer引物进行pcr,纯化pcr产物;
(5)取300ngpcr产物与1ul10xt7eibuffer混匀,按以下pcr程序进行复性95℃10min,95℃至85℃-2℃/s,85℃到25℃-0.25℃/s,25℃持续1min。
(6)复性之后产物加入1ult7ei,37℃酶切20min,跑2%琼脂糖胶,结果如图7所示,采用灰度分析可得对trac、trbc、b2m、ctla4四个基因靶点的切割效率分别为15%,25%,45%,45%。
实施例5
(1)选取dnmt1、hbb、il12a、polq、b2m5个基因5个靶点进行脱靶分析,用软件(http://www.rgenome.net/cas-offinder/)预测了41个脱靶位点,并设计相应引物。
(2)24孔板共转as/lb/cecas12a真核表达质粒(700ng)和u6-crrnaspacer真核表达质粒(300ng),48h后裂解细胞,取1ul裂解液作为模板并采用1)中设计的引物进行扩增,然后纯化,进行二代测序,统计结构如图8所示,ascas12a,lbcas12a,cecas12a都具有很低的脱靶率,但cecas12a相比于ascas12a,lbcas12a具有更低的脱靶率。
显然,上述实施例仅仅是为清楚地说明所作的实例,而并非对实施方式的限制。对于所属领域的普通技术人员来说,在上述说明的基础上还可以做出其它不同形式的变化或变动。这里无需也无法对所有的实施方式予以穷举。而因此所引申的显而易见的变化或变动仍处于本发明创造的保护范围之内。
序列表
<110>武汉大学
<120>ii类v型crispr蛋白cecas12a及其用于基因编辑的应用
<160>2
<210>1
<211>1286
<212>prt
<213>coprococcuseutactus细菌
<400>1
asnasnasnthrasnasnserpheglupropheileglyglyasnser
151015
valserlysthrleuargasngluleuargvalglyserglutyrthr
202530
glylyshisilelysglucysalaileilealagluaspalavallys
354045
alagluasnglntyrilevallysglumetmetaspaspphetyrarg
505560
asppheileasnarglysleuaspalaleuglnglyileasntrpglu
65707580
glnleupheaspilemetlyslysalalysleuasplysserasnlys
859095
valserlysgluleuasplysileglngluserthrarglysgluile
100105110
vallysilepheserseraspproiletyrlysaspmetleulysala
115120125
aspmetileserlysileleuproglutyrilevalasplystyrgly
130135140
aspalaalaserargileglualavallysvalphetyrglypheser
145150155160
glytyrpheileaspphetrpalaserarglysasnvalpheserasp
165170175
lysasnilealaseralaileprohisargilevalasnvalasnala
180185190
argilehisleuaspasnilethralapheasnargilealagluile
195200205
alaglyaspgluvalalaglyilealagluaspalacysalatyrleu
210215220
glnasnmetserleugluaspvalphethrglyalacystyrglyglu
225230235240
pheilecysglnlysaspileaspargtyrasnasnilecysglyval
245250255
ileasnglnhismetasnglntyrcysglnasnlyslysileserarg
260265270
serlysphelysmetgluargleuhislysglnileleucysargser
275280285
gluserglyphegluileproileglypheglnthraspglygluval
290295300
ileaspalaileasnserpheserthrileleugluglulysaspile
305310315320
leuaspargleuargthrleuserglngluvalthrglytyraspmet
325330335
gluargiletyrvalserserlysalaphegluservalserlystyr
340345350
ileasphislystrpaspvalilealasersermettyrasntyrphe
355360365
serglyalavalargglylysaspasplyslysaspalalysilegln
370375380
thrgluilelyslysilelyssercysserleuleuaspleulyslys
385390395400
leuvalaspmettyrtyrlysmetaspglymetcysleugluhisglu
405410415
alathrglutyrvalalaglyilethrgluileleuvalasppheasn
420425430
tyrlysthrpheaspmetaspaspservallysmetileglnasnglu
435440445
hismetileasngluilelysglutyrleuaspthrtyrmetserile
450455460
tyrhistrpalalysaspphemetileaspgluleuvalaspargasp
465470475480
metgluphetyrsergluleuaspgluiletyrtyraspleuserasp
485490495
ilevalproleutyrasnlysvalargasntyrvalthrglnlyspro
500505510
tyrserglnasplysilelysleuasnpheglyserprothrleuala
515520525
asnglytrpserlysserlysglupheaspasnasnvalvalvalleu
530535540
leuargaspglulysiletyrleualaileleuasnvalglyasnlys
545550555560
proserlysaspilemetalaglygluaspargargargseraspthr
565570575
asptyrlyslysmetasntyrtyrleuleuproglyalaserlysthr
580585590
leuprohisvalpheileserserasnalatrplyslysserhisgly
595600605
ileproaspgluilemettyrglytyrasnglnasnlyshisleulys
610615620
serserproasnpheaspleugluphecysarglysleuileasptyr
625630635640
tyrlysglucysileaspsertyrproasntyrglnilepheasnphe
645650655
lysphealaalathrgluthrtyrasnaspilesergluphetyrlys
660665670
aspvalgluargglnglytyrlysileglutrpsertyrileserglu
675680685
aspaspileasnglnmetaspargaspglyglniletyrleuphegln
690695700
iletyrasnlysaspphealaproasnserlysglymetglnasnleu
705710715720
histhrleutyrleulysasnilepheserglugluasnleuserasp
725730735
valvalilelysleuasnglyglualagluleuphephearglysser
740745750
serileglnhislysargglyhislyslysglyservalleuvalasn
755760765
lysthrtyrlysthrthrglulysthrgluasnglyglnglygluile
770775780
gluvalilegluservalproaspglncystyrleugluleuvallys
785790795800
tyrtrpsergluglyglyvalglyglnleusergluglualaserlys
805810815
tyrlysasplysvalserhistyralaalathrmetaspilevallys
820825830
aspargargtyrthrgluasplysphepheilehismetproilethr
835840845
ileasnphelysalaaspasnargasnasnvalasnglulysvalleu
850855860
lyspheilealagluasnaspaspleuhisvalileglyileasparg
865870875880
glygluargasnleuleutyrvalservalileaspserargglyarg
885890895
ilevalgluglnlysserpheasnilevalgluasntyrgluserser
900905910
lysasnvalileargarghisasptyrlysglylysleuvalasnlys
915920925
gluhistyrargasnglualaarglyssertrplysgluileglylys
930935940
ilelysgluilelysgluglytyrleuserglnvalilehisgluile
945950955960
serlysleuvalleulystyrasnalaileilevalmetgluaspleu
965970975
asntyrglyphelysargglyargphelysvalgluargglnvaltyr
980985990
glnlysphegluthrmetleuileasnlysleualatyrleuvalasp
99510001005
lysserargalavalaspgluproglyglyleuleulysglytyrgln
101010151020
leuthrtyrvalproaspasnleuglygluleuglyserglncysgly
1025103010351040
ileilephetyrvalproalaalatyrthrserlysileaspproval
104510501055
thrglyphevalaspvalpheaspphelysalatyrserasnalaglu
106010651070
alaargleuasppheileasnlysleuaspcysileargtyraspala
107510801085
serargasnlysphegluilealapheasptyrglyasnpheargthr
109010951100
hishisthrthrleualalysthrsertrpthrilepheilehisgly
1105111011151120
aspargilelyslysgluargglysertyrglytrplysaspgluile
112511301135
ileaspileglualaargilearglysleuphegluaspthraspile
114011451150
glutyralaaspglyhisasnleuileglyaspileasngluleuglu
115511601165
serproileglnlyslysphevalglygluleupheaspileilearg
117011751180
phethrvalglnleuargasnserlysserglulystyraspglythr
1185119011951200
glulysglutyrasplysileileserprovalmetaspgluglugly
120512101215
valphephethrthraspsertyrileargalaaspglythrgluleu
122012251230
prolysaspalaaspalaasnglyalatyrcysilealaleulysgly
123512401245
leutyraspvalleualavallyslystyrtrplysgluglyglulys
125012551260
pheasparglysleuleualailethrasntyrasntrppheaspphe
1265127012751280
ileglnasnargargphe
1285
<210>2
<211>3858bp
<212>dna
<213>coprococcuseutactus细菌
<400>2
aacaacaacaccaacaacagcttcgagcccttcatcggcggcaacagcgtgagcaagacc60
ctgcgcaacgagctgcgcgtgggcagcgagtacaccggcaagcacatcaaggagtgcgcc120
atcatcgccgaggacgccgtgaaggccgagaaccagtacatcgtgaaggagatgatggac180
gacttctaccgcgacttcatcaaccgcaagctggacgccctgcagggcatcaactgggag240
cagctgttcgacatcatgaagaaggccaagctggacaagagcaacaaggtgagcaaggag300
ctggacaagatccaggagagcacccgcaaggagatcgtgaagatcttcagcagcgacccc360
atctacaaggacatgctgaaggccgacatgatcagcaagatcctgcccgagtacatcgtg420
gacaagtacggcgacgccgccagccgcatcgaggccgtgaaggtgttctacggcttcagc480
ggctacttcatcgacttctgggccagccgcaagaacgtgttcagcgacaagaacatcgcc540
agcgccatcccccaccgcatcgtgaacgtgaacgcccgcatccacctggacaacatcacc600
gccttcaaccgcatcgccgagatcgccggcgacgaggtggccggcatcgccgaggacgcc660
tgcgcctacctgcagaacatgagcctggaggacgtgttcaccggcgcctgctacggcgag720
ttcatctgccagaaggacatcgaccgctacaacaacatctgcggcgtgatcaaccagcac780
atgaaccagtactgccagaacaagaagatcagccgcagcaagttcaagatggagcgcctg840
cacaagcagatcctgtgccgcagcgagagcggcttcgagatccccatcggcttccagacc900
gacggcgaggtgatcgacgccatcaacagcttcagcaccatcctggaggagaaggacatc960
ctggaccgcctgcgcaccctgagccaggaggtgaccggctacgacatggagcgcatctac1020
gtgagcagcaaggccttcgagagcgtgagcaagtacatcgaccacaagtgggacgtgatc1080
gccagcagcatgtacaactacttcagcggcgccgtgcgcggcaaggacgacaagaaggac1140
gccaagatccagaccgagatcaagaagatcaagagctgcagcctgctggacctgaagaag1200
ctggtggacatgtactacaagatggacggcatgtgcctggagcacgaggccaccgagtac1260
gtggccggcatcaccgagatcctggtggacttcaactacaagaccttcgacatggacgac1320
agcgtgaagatgatccagaacgagcacatgatcaacgagatcaaggagtacctggacacc1380
tacatgagcatctaccactgggccaaggacttcatgatcgacgagctggtggaccgcgac1440
atggagttctacagcgagctggacgagatctactacgacctgagcgacatcgtgcccctg1500
tacaacaaggtgcgcaactacgtgacccagaagccctacagccaggacaagatcaagctg1560
aacttcggcagccccaccctggccaacggctggagcaagagcaaggagttcgacaacaac1620
gtggtggtgctgctgcgcgacgagaagatctacctggccatcctgaacgtgggcaacaag1680
cccagcaaggacatcatggccggcgaggaccgccgccgcagcgacaccgactacaagaag1740
atgaactactacctgctgcccggcgccagcaagaccctgccccacgtgttcatcagcagc1800
aacgcctggaagaagagccacggcatccccgacgagatcatgtacggctacaaccagaac1860
aagcacctgaagagcagccccaacttcgacctggagttctgccgcaagctgatcgactac1920
tacaaggagtgcatcgacagctaccccaactaccagatcttcaacttcaagttcgccgcc1980
accgagacctacaacgacatcagcgagttctacaaggacgtggagcgccagggctacaag2040
atcgagtggagctacatcagcgaggacgacatcaaccagatggaccgcgacggccagatc2100
tacctgttccagatctacaacaaggacttcgcccccaacagcaagggcatgcagaacctg2160
cacaccctgtacctgaagaacatcttcagcgaggagaacctgagcgacgtggtgatcaag2220
ctgaacggcgaggccgagctgttcttccgcaagagcagcatccagcacaagcgcggccac2280
aagaagggcagcgtgctggtgaacaagacctacaagaccaccgagaagaccgagaacggc2340
cagggcgagatcgaggtgatcgagagcgtgcccgaccagtgctacctggagctggtgaag2400
tactggagcgagggcggcgtgggccagctgagcgaggaggccagcaagtacaaggacaag2460
gtgagccactacgccgccaccatggacatcgtgaaggaccgccgctacaccgaggacaag2520
ttcttcatccacatgcccatcaccatcaacttcaaggccgacaaccgcaacaacgtgaac2580
gagaaggtgctgaagttcatcgccgagaacgacgacctgcacgtgatcggcatcgaccgc2640
ggcgagcgcaacctgctgtacgtgagcgtgatcgacagccgcggccgcatcgtggagcag2700
aagagcttcaacatcgtggagaactacgagagcagcaagaacgtgatccgccgccacgac2760
tacaagggcaagctggtgaacaaggagcactaccgcaacgaggcccgcaagagctggaag2820
gagatcggcaagatcaaggagatcaaggagggctacctgagccaggtgatccacgagatc2880
agcaagctggtgctgaagtacaacgccatcatcgtgatggaggacctgaactacggcttc2940
aagcgcggccgcttcaaggtggagcgccaggtgtaccagaagttcgagaccatgctgatc3000
aacaagctggcctacctggtggacaagagccgcgccgtggacgagcccggcggcctgctg3060
aagggctaccagctgacctacgtgcccgacaacctgggcgagctgggcagccagtgcggc3120
atcatcttctacgtgcccgccgcctacaccagcaagatcgaccccgtgaccggcttcgtg3180
gacgtgttcgacttcaaggcctacagcaacgccgaggcccgcctggacttcatcaacaag3240
ctggactgcatccgctacgacgccagccgcaacaagttcgagatcgccttcgactacggc3300
aacttccgcacccaccacaccaccctggccaagaccagctggaccatcttcatccacggc3360
gaccgcatcaagaaggagcgcggcagctacggctggaaggacgagatcatcgacatcgag3420
gcccgcatccgcaagctgttcgaggacaccgacatcgagtacgccgacggccacaacctg3480
atcggcgacatcaacgagctggagagccccatccagaagaagttcgtgggcgagctgttc3540
gacatcatccgcttcaccgtgcagctgcgcaacagcaagagcgagaagtacgacggcacc3600
gagaaggagtacgacaagatcatcagccccgtgatggacgaggagggcgtgttcttcacc3660
accgacagctacatccgcgccgacggcaccgagctgcccaaggacgccgacgccaacggc3720
gcctactgcatcgccctgaagggcctgtacgacgtgctggccgtgaagaagtactggaag3780
gagggcgagaagttcgaccgcaagctgctggccatcaccaactacaactggttcgacttc3840
atccagaaccgccgcttc3858
- 还没有人留言评论。精彩留言会获得点赞!